Protein-based therapeutics, like monoclonal antibodies (mAbs) and antibody-drug conjugates (ADCs), have revolutionized the pharmaceutical industry, offering advantages over traditional small molecule drugs, such as high affinity, potency, and specificity, with low toxicity and minimal adverse effects. However, challenges related to protein folding, purification, stability, and immunogenicity present hurdles in the development and manufacturing processes of these therapeutics.
Protein stability throughout the manufacturing, storage, and delivery process is essential, as structural instability resulting from misfolding, unfolding, modifications, and aggregation poses a risk to the efficacy of these drugs, overshadowing their attributes. Protein aggregation can create formidable challenges in therapeutic protein development, undermining efficacy, product quality, and patient safety 4. Thus addressing aggregation propensity is imperative in enabling the development and commercialization of clinically and commercially viable protein therapeutics. Insights from recent research on therapeutic protein aggregation can help mitigate the challenges by outlining holistic strategies for aggregation control and prevention.
Protein aggregation
Fundamentally, aggregation starts when disturbances destabilize proteins, making them unfold partially. Elucidating the molecular stages comprising the aggregation cascade is imperative to guide targeted strategies that control aggregate levels. Recent research from Rahban et. al conceptualizes a simplified yet insightful framework that captures the sequence of events driving aggregation mediated by destabilized folding intermediates2:
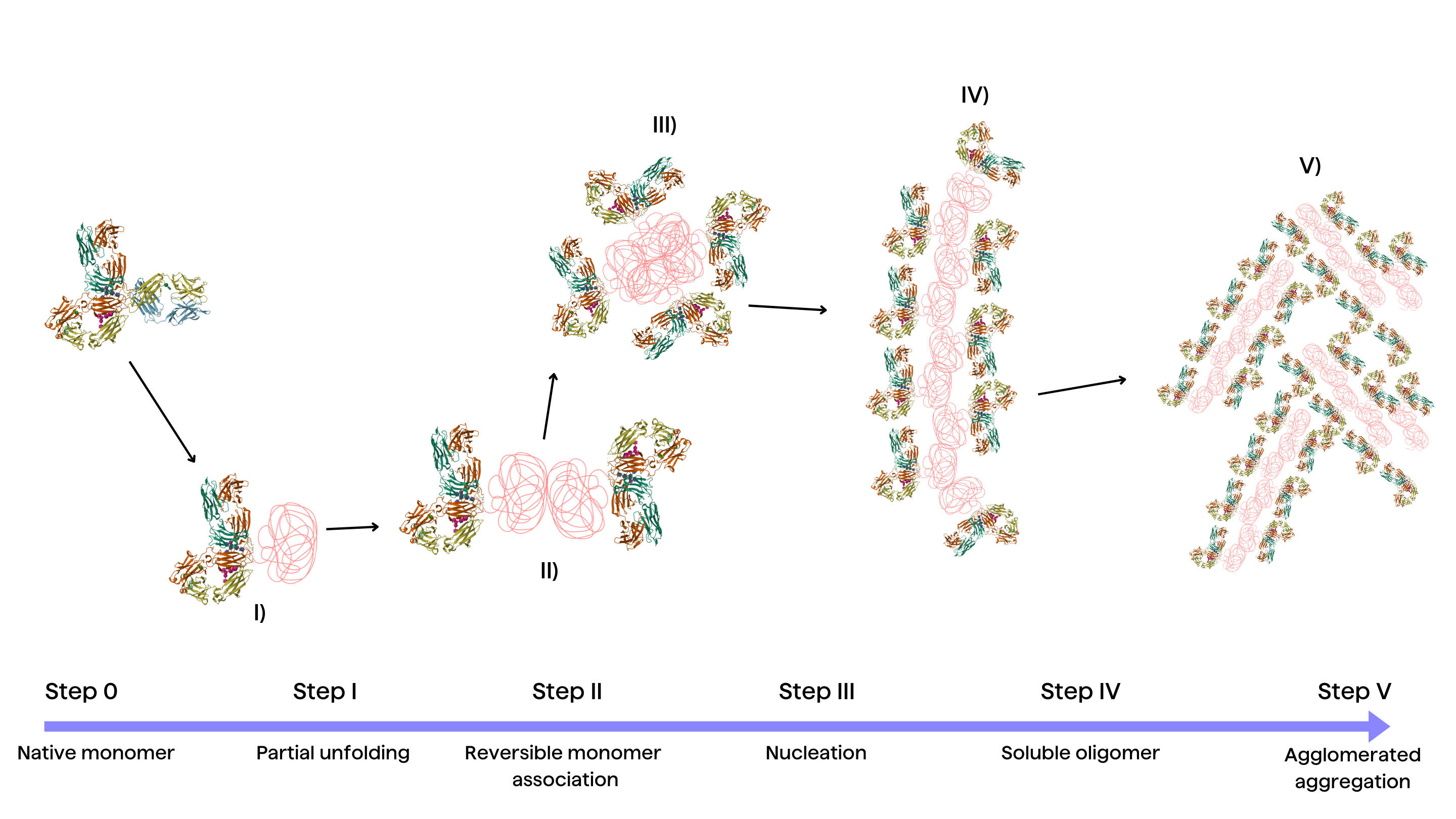
Stage I: Partial unfolding
Partial protein unfolding, caused by environmental factors like temperature, is the base condition for aggregation-prone conformations3. The energy barriers limiting transient structural fluctuations and unfolding need to be judiciously tuned.
Stage II: Reversible monomer association
Unfolded monomers associate via hydrophobic interactions and hydrogen bonding. Electrostatic repulsions can however limit non-native intermolecular contacts22.
Stage III: Nucleation
The rate-limiting nucleation phase involves structural reorganization into aggregated nuclei enriched in β-sheets, setting the stage for rapid aggregate elongation21.
Stage IV: Growth by monomer addition
Monomers rapidly accrete onto aggregation nuclei, accelerating aggregate growth3. Preformed aggregates can also disrupt protein homeostasis, further propagating aggregation.
Stage V: Aggregate association
Soluble high molecular weight aggregates accumulate, which may ultimately precipitate out of solution depending on solubility limits3.
Drivers and consequences of aggregation
Proteins have an intrinsic tendency to aggregate, especially when destabilized from their native conformations 5. Several factors drive the dynamics and pathways of therapeutic protein aggregation
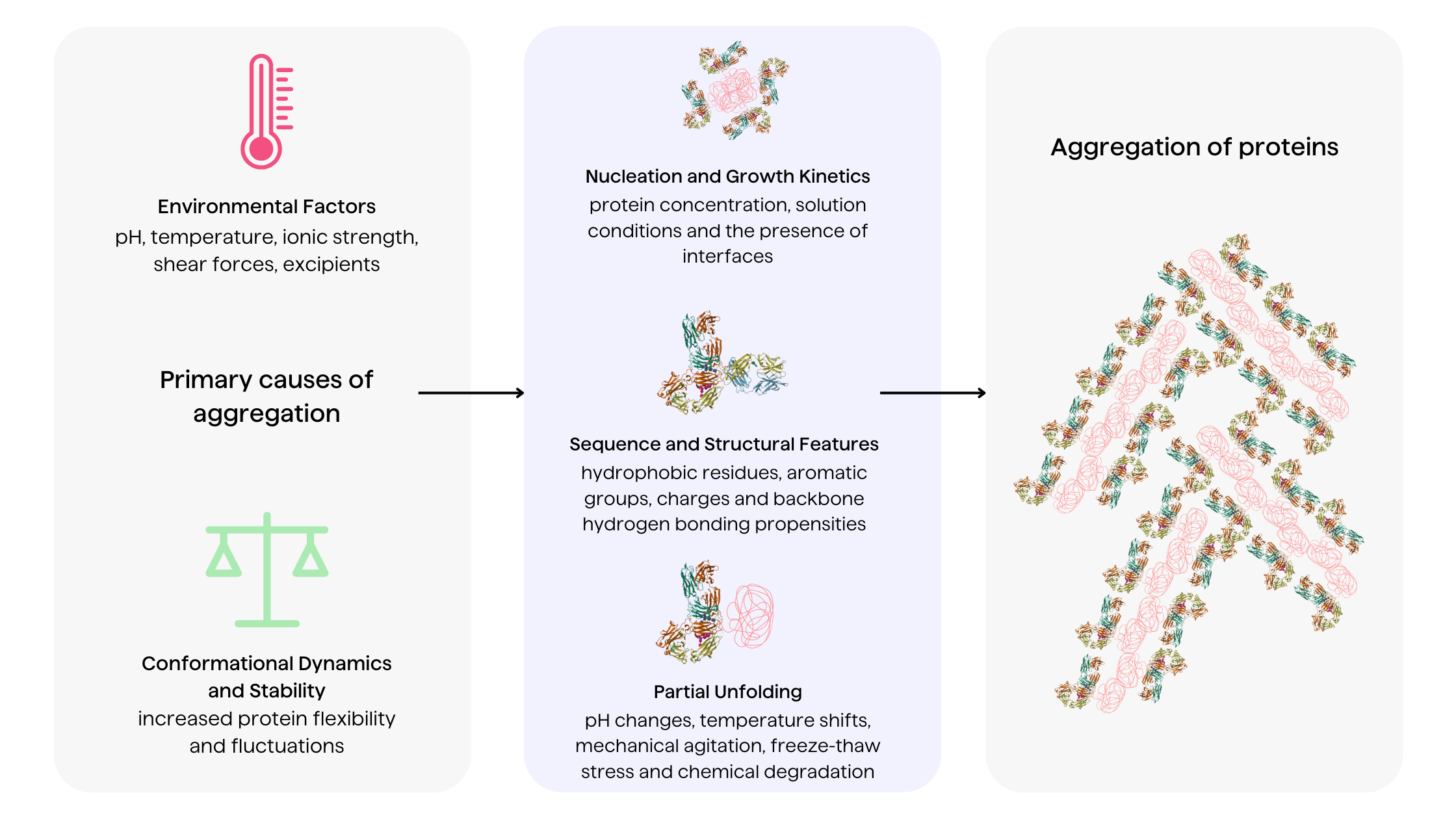
Environmental factors
Solution conditions including pH, temperature, ionic strength, shear forces and boundaries between states (liquid, gas, solid) profoundly impact aggregation pathways 1,19. Extrinsic factors modulate protein conformation, influencing the exposure of intermolecular interactions. Excipients like salts, surfactants and polyols also affect aggregation mechanisms through preferential interactions8.
Amino acid sequence and structural features
The distribution of hydrophobic residues, aromatic groups, charges and backbone hydrogen bonding propensities encoded within the amino acid sequence dictates localized propensities 1,20. Key antibody regions like the CDRs, VH-VL interface and framework regions harbor sequence and structural vulnerabilities predisposing proteins to aggregation3.
Partial unfolding of proteins
Partial unfolding of proteins constitutes a key trigger that enhances their aggregation susceptibility by increasing solvent exposure of aggregation-prone regions 1,17. Destabilized proteins are prone to self-association between exposed hydrophobic patches, leading to nucleation and aggregate growth 1,2. Factors causing transient unfolding include pH changes, temperature shifts, mechanical agitation, freeze-thaw stress and chemical degradation12.
Nucleation and growth kinetics
Protein aggregation displays complex nucleation-dependent polymerization kinetics. Following an initial lag nucleation phase, aggregates grow rapidly by monomer addition and aggregate fusion 6. The kinetics are influenced by protein concentration, solution conditions and the presence of interfaces that catalyze aggregation7. Preformed aggregates can also disrupt protein homeostasis, propagating further aggregation.
Conformational dynamics and stability
The delicate balance between conformational stability and dynamics governs aggregation risks. Increased protein flexibility and fluctuations augment the population of destabilized aggregation-prone species whereas excessive rigidity also induces aggregation through decreased solubility18. Optimizing stability while retaining sufficient dynamics is key for functionality.
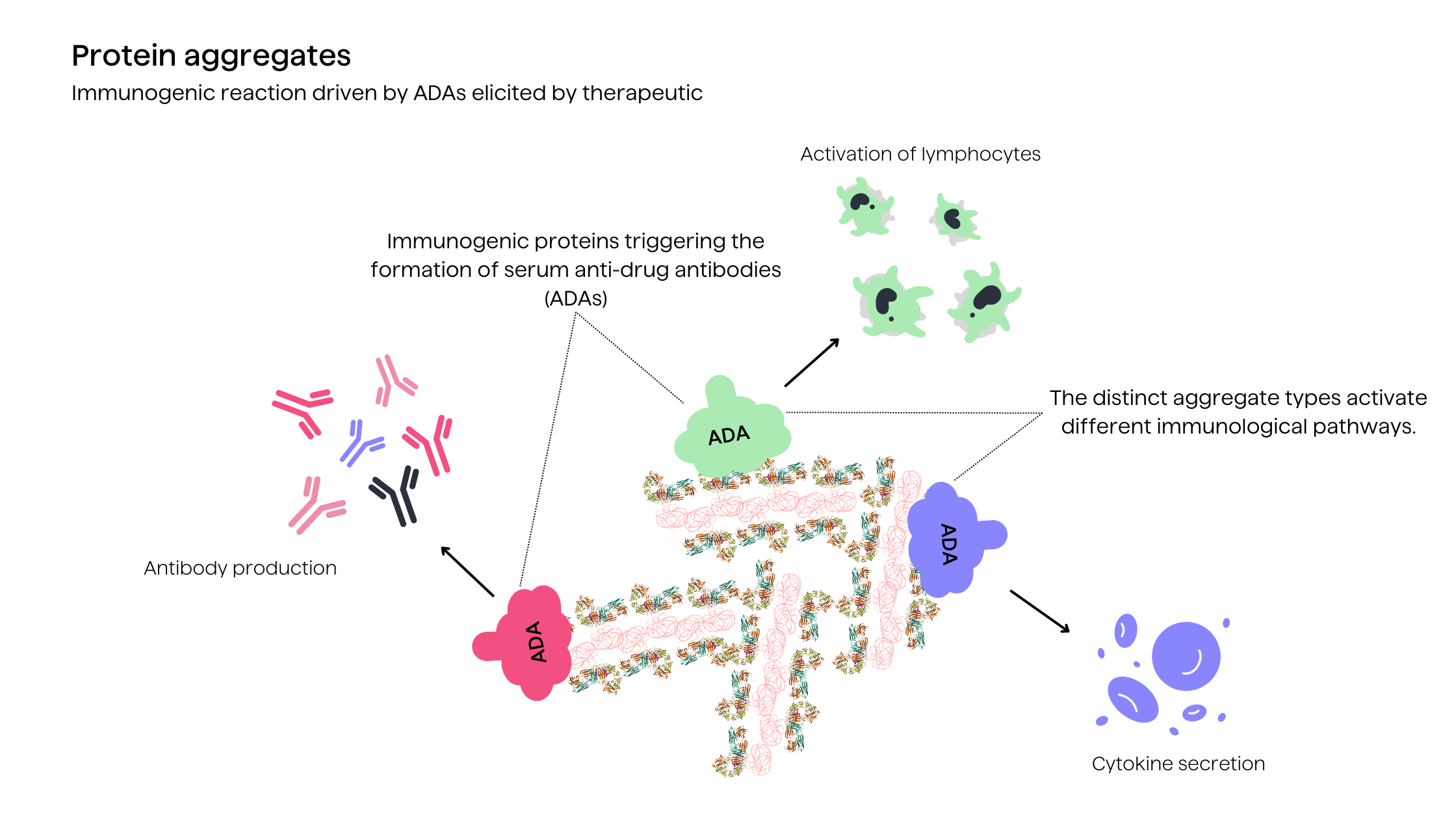
As for major safety concerns associated with aggregates, it is their increased immunogenic potential which can provoke drug-neutralizing immune responses or cause adverse hypersensitivity reactions11. The altered structural conformations of aggregates promote recognition as foreign antigens by the immune system2. Aggregates potentially disrupt immune tolerance established towards the therapeutic protein (in this case antibody drug).
Furthermore, protein degradation products and impurities in formulations may also contribute to unwanted immunogenicity2. Computational immunogenicity prediction platforms have emerged to predict such risks and guide protein engineering efforts to deimmunize aggregation-prone regions3. Overall, managing aggregation is imperative both for preserving efficacy and mitigating the potential safety risks caused by increased immune response.
Approaches for mitigating protein aggregation
Comprehensive protein engineering approaches, synergized by rationally designed formulation and process conditions, can effectively mitigate aggregation risks in biopharmaceutical development9.
Protein Engineering
Strategies include improving conformational stability through structure-guided mutagenesis, modulating interfacial dynamics by altering surface properties, residues at protein-protein interfaces and introduction of repulsive electrostatic interactions 10,13.
Formulation Optimization
Excipients modulate solution behavior and inhibit aggregation through preferential exclusion mechanisms that promote protein hydration and inhibit nonspecific interactions2. Stabilizers like arginine suppress aggregation by impeding protein-protein contacts and interfering with hydrophobic interactions.
Manufacturing Process Optimization
Strategies encompass eliminating mechanical stresses through low-shear processing, minimizing air-water interfacial stresses via exclusion of gas-liquid interfaces and optimizing purification procedures to remove aggregates and other impurities14. Inline monitoring tools help control process parameters.
Storage Condition Optimization
Aggregation kinetics display high temperature-sensitivity, necessitating stringent cold chain maintenance. Suitable packaging prevents ingress of moisture and oxygen. Light protection and freeze-thaw avoidance are also critical2.
Role of computational techniques
Modern computational methods
Computation methods offer invaluable predictive capabilities to gain molecular insights into protein aggregation while guiding experimental efforts to design aggregation-resistant proteins1.
Molecular simulations techniques
Techniques like molecular dynamics simulations investigate protein dynamics at atomic resolution, predicting regions that are prone to aggregation. Binding free energy calculations assess protein-protein interaction propensities15.
Structure-based protein engineering computational tools
Protein design tools facilitate the strategic introduction of mutations predicted to modulate stability or interfere with aggregation-prone conformers while retaining function.2
Machine learning solutions
Supervised learning algorithms leverage protein sequence and physicochemical descriptors to predict aggregation propensity, classify aggregates and guide sequence optimization approaches to suppress aggregation16.
De-risking development of therapeutics
Therapeutic protein aggregation undermines biopharmaceutical quality, efficacy and safety, necessitating substantial efforts to mitigate risks and design aggregation-resistant candidates. Strategies require comprehensive biophysical understanding of molecular triggering factors, nucleation phenomena, growth pathways and environmental modulation of aggregation coupled with state-of-the-art computational methodologies for predictive analysis and design.
Additionally, the immunological implications pose imperative considerations during candidate selection and process development to minimize clinical risks. Holistic solutions spanning discovery, formulation sciences and manufacturing process optimization can effectively address aggregation issues in biopharmaceutical development to enable the successful translation of promising candidates.








